Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCι in complex with a substrate peptide from Par-3
2012.05.09Wang, C., Shang, Y., Yu, J., & Zhang, M. (2012).Structure, 20(5), 791-801.
Protein kinase C (PKC) play critical roles in many cellular functions including differentiation, proliferation, growth, and survival. However, the molecular bases governing PKC's substrate recognitions remain poorly understood. Here we determined the structure of PKCι in complex with a peptide from Par-3 at 2.4 Å. PKCι in the complex adopts catalytically competent, closed conformation without phosphorylation of Thr402 in the activation loop. The Par-3 peptide binds to an elongated groove formed by the N- and C-lobes of the kinase domain. The PKCι/Par-3 complex structure, together with extensive biochemical studies, reveals a set of substrate recognition sites common to all PKC isozymes as well as a hydrophobic pocket unique to aPKC. A consensus aPKC's substrate recognition sequence pattern can be readily identified based on the complex structure. Finally, we demonstrate that the pseudosubstrate sequence of PKCι resembles its substrate sequence, directly binds to and inhibits the activity of the kinase.
- Recommend
-
2025-10-22
IQSEC2/BRAG1 may modulate postsynaptic density assembly through Ca2+-induced phase separation.
-
2025-08-22
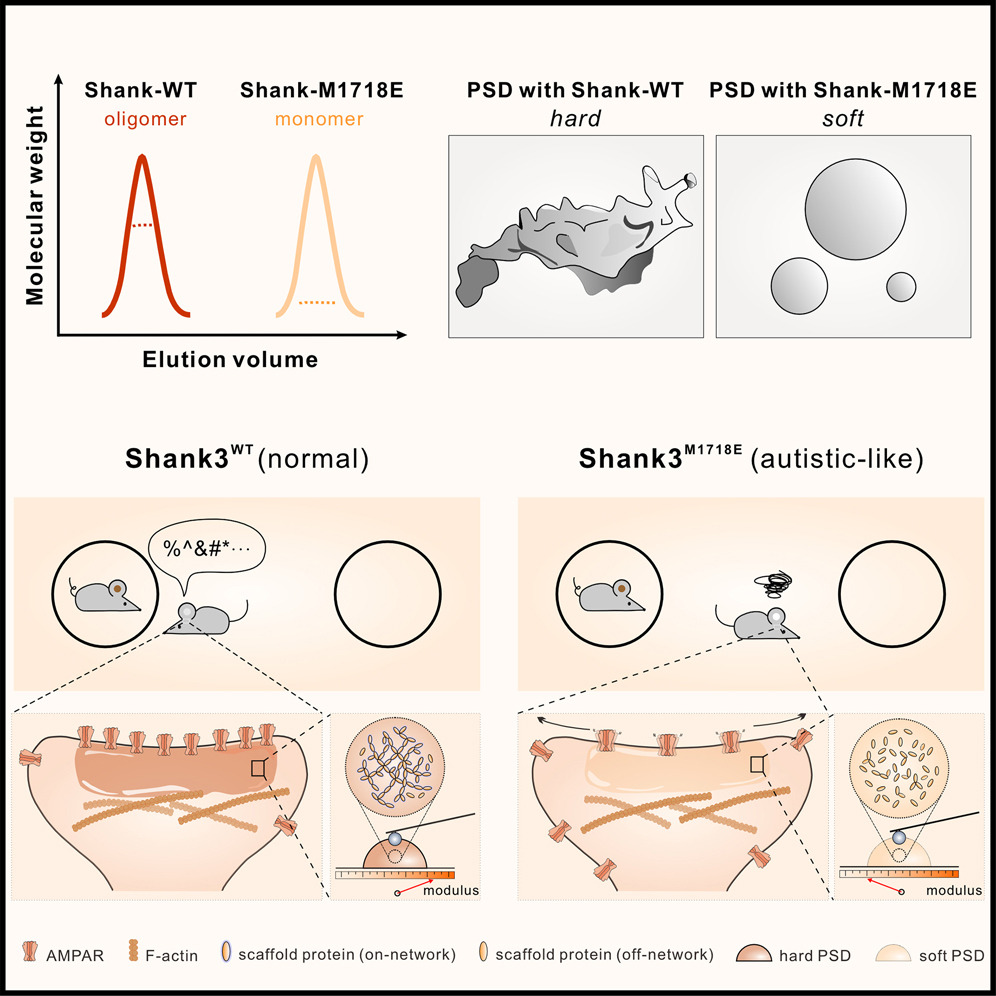
Shank3 oligomerization governs material properties of the postsynaptic density condensate and synaptic plasticity.
-
2025-08-21
Modulating synaptic glutamate receptors by targeting network nodes of the postsynaptic density condensate.
-
2025-08-19
Current practices in the study of biomolecular condensates: a community comment.
-
2025-06-10
Phase separation instead of binding strength determines target specificities of MAGUKs.