Substrate docking–mediated specific and efficient lysine methylation by the SET domain–containing histone methyltransferase SETD7
2019.07.19Liu, H., Li, Z., Yang, Q., Liu, W., Wan, J., Li, J., & Zhang, M. (2019). Journal of Biological Chemistry, 294(36), 13355-13365.
Lysine methylation of cellular proteins is catalyzed by dozens of lysine methyltransferases (KMTs), occurs in thousands of different histone and nonhistone proteins, and regulates diverse biological processes. Dysregulation of KMT-mediated lysine methylations underlies many human diseases. A key unanswered question is how proteins, nonhistone proteins in particular, are specifically methylated by each KMT. Here, using several biochemical approaches, including analytical gel filtration chromatography, isothermal titration calorimetry, and in vitro methylation assays, we discovered that SET domain–containing 7 histone lysine methyltransferase (SETD7), a KMT capable of methylating both histone and nonhistone proteins, uses its N-terminal membrane occupation and recognition nexus (MORN) repeats to dock its substrates and subsequently juxtapose their Lys methylation motif for efficient and specific methylation by the catalytic SET domain. Such docking site–mediated methylation mechanism rationalizes binding and methylation of previously known substrates and predicts new SETD7 substrates. Our findings further suggest that other KMTs may also use docking-mediated substrate recognition mechanisms to achieve their catalytic specificity and efficiency.
- Recommend
-
2025-10-22
IQSEC2/BRAG1 may modulate postsynaptic density assembly through Ca2+-induced phase separation.
-
2025-08-22
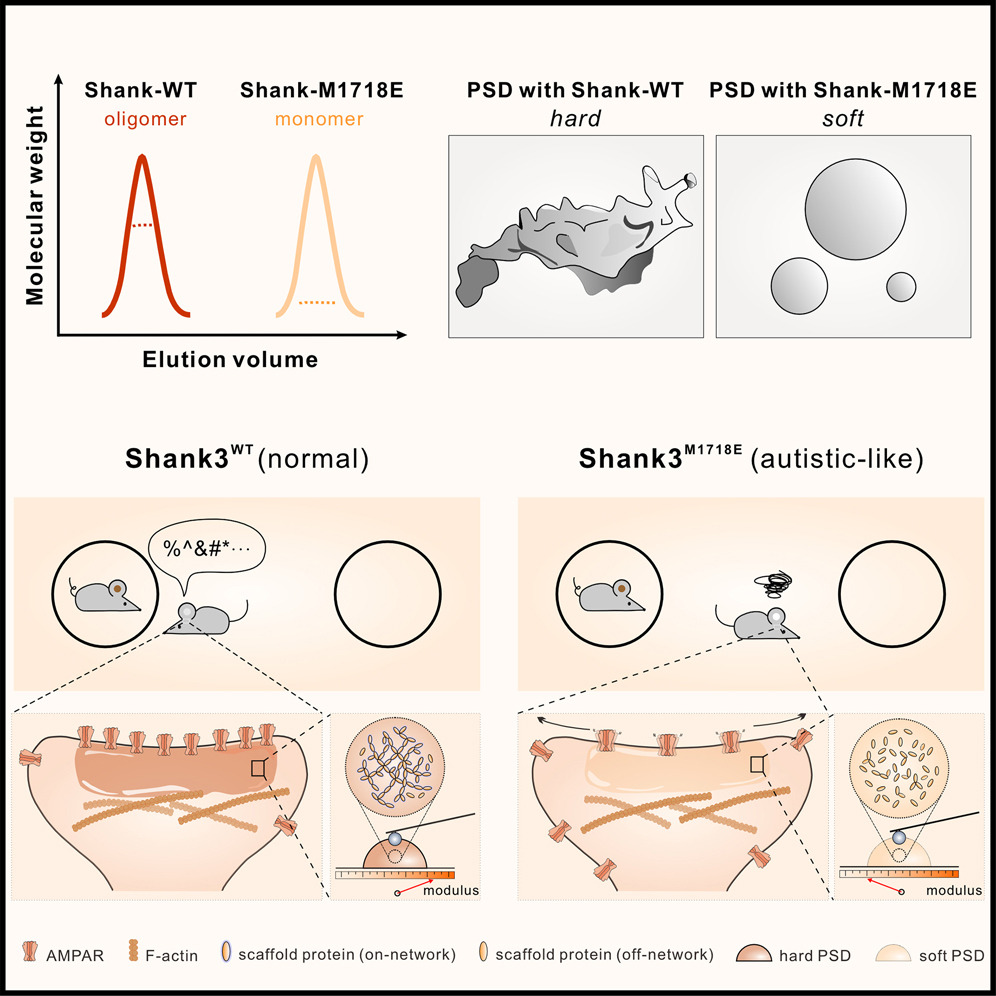
Shank3 oligomerization governs material properties of the postsynaptic density condensate and synaptic plasticity.
-
2025-08-21
Modulating synaptic glutamate receptors by targeting network nodes of the postsynaptic density condensate.
-
2025-08-19
Current practices in the study of biomolecular condensates: a community comment.
-
2025-06-10
Phase separation instead of binding strength determines target specificities of MAGUKs.